

The fingerprints of bacteria
We don’t just eat livestock. We live in close proximity, and because some bacterial infections can harm both humans and livestock, our health is interconnected. An example is the outbreak of the MERS virus in the Middle East that began in camels and was transferred to humans, killing at least 100 humans by the end of April 2014. The Saudi Arabian government urged its people to wear face masks and gloves around camels to prevent the spread of the virus, but the idea that cattle could sicken humans seemed absurd to many of its citizens. Camel owners began posting pictures of themselves kissing camels to social media to show they did not believe the government’s claim, an act that some of them may later regret.(A2)

Another example, one closer to home, is the bacteria Staphylococcus aureus, or S. aureus, for short. These bacteria are everywhere: on our skin and in our nose. Most do no harm , but just as their are different ethnicities of humans there are different strains of S. aureus, and some threaten both cattle and humans.
Figure 1—S. aureus, as seen by a scanning electron micrograph
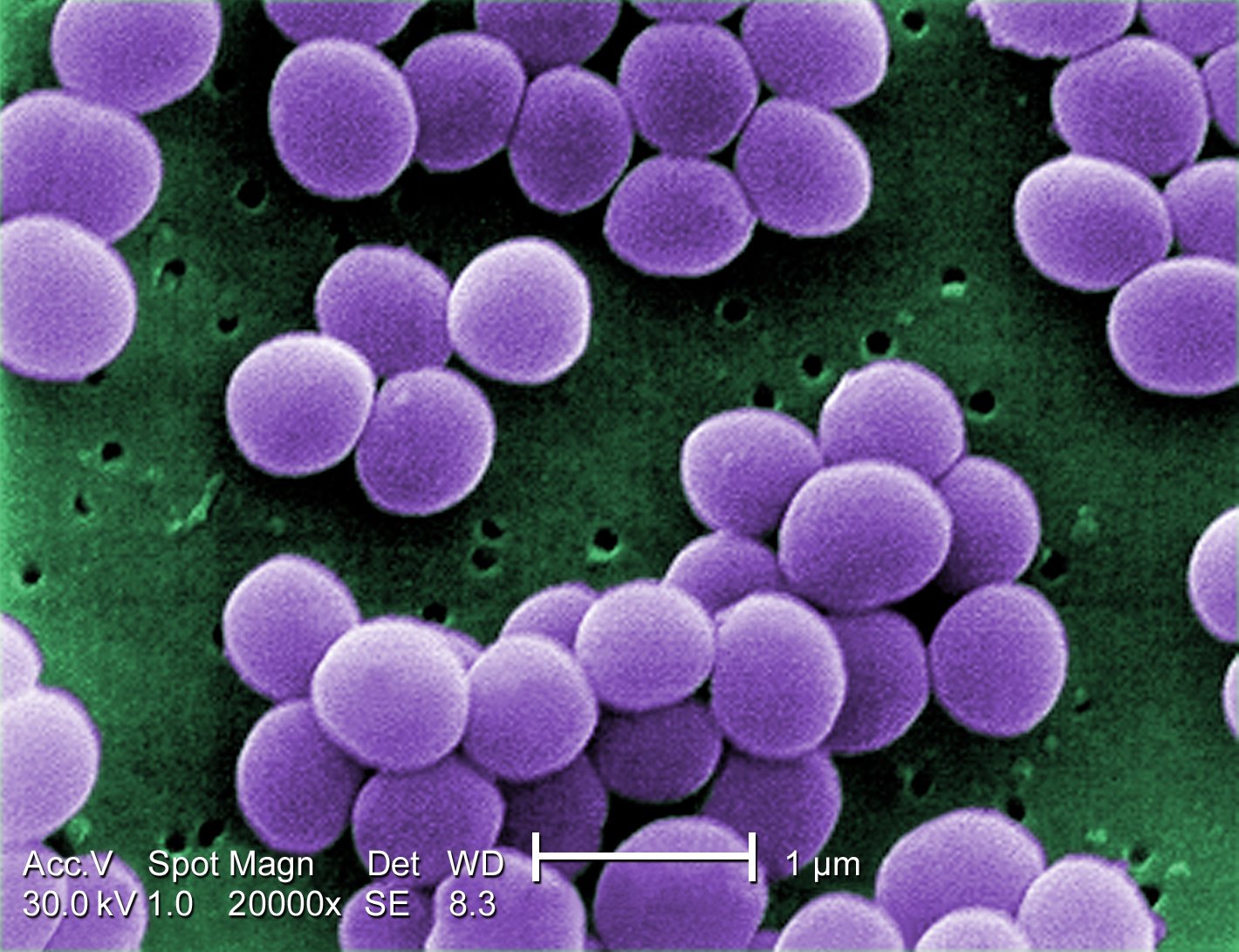
When S. aureus infects the udders of dairy cattle or sheep they can cause a painful infection referred to as mastitis. Often antibiotics like methicillin will cure the cow, but sometimes there is no remedy. This strain of bacteria can also infect humans, and because some have become resistant to antibiotics they can pose considerably harm, even death. These resistant strains are referred to as methicillin-resistant Staphylococcus aureus. It’s acronym you may be familiar with: MRSA.
Figure 2—MRSA infection on skin of human
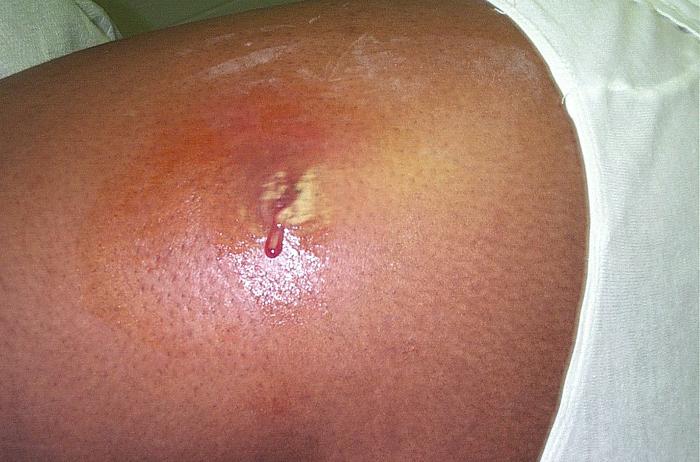
To what extent can MRSA in cattle infect humans? To what extent does MRSA infections in humans spread to cattle? These are important questions. Protecting human health from antibiotic-resistant bacteria requires us to understand the linkages between the health of humans and cattle. To what extent is the MRSA in cattle the same MRSA that infect humans? This lecture is all about answering that question, and we will do so by taking a tour of the laboratories at the Department of Biochemistry and Molecular Biology at Oklahoma State University to witness the methods employed in a recent study published in the Journal of Dairy Science(M1), where Dr. Stephanie Matyi and her associates showed that the same strain of bacteria infected cattle at both hospitals and dairy farms in New Mexico. This study entailed gathering samples of milk from dairy cattle, determining whether any S. aureus were present and whether they were the same S. aureus found in hospitals. It required the researchers to go deep within this tiny organism to study something even tinier: its DNA.
Figure 3—Severe mastitis infection in sheep
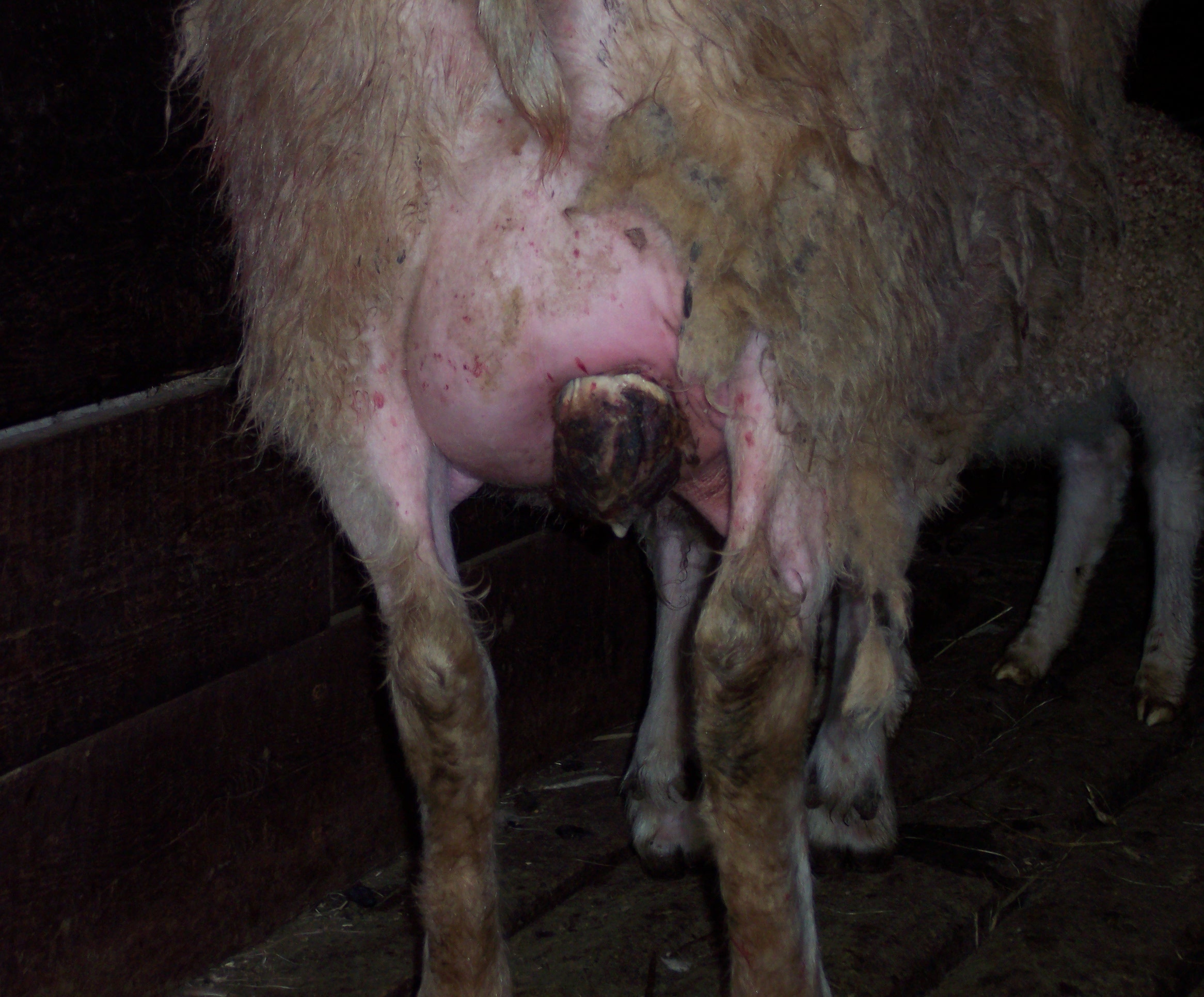
Identifying MRSA
Step 1 in the Matyi study: Acquire milk samples
Dr. Matyi and her associates already knew the genetic makeup of MRSA in hospitals, so she needed to gather samples of milk to determine whether the milk contained the S. aureus. A total of 133 milk samples were gathered from 133 different cows. Most of these cattle were healthy, but 33 were sick, so it was likely that some bacterial infection of some kind would be confirmed. The question was whether any of those bacteria were S. aureus.
Step 2 in Matyi study: Feed S. aureus, poison others
Suppose you wanted to identify which students in a dormitory were meat-eaters, and suppose you could not communicate with them. You could place them in a cafeteria for six hours with nothing to eat but meat. This setting has food for the meat-eaters but no food to nourish vegetarians. It would not take long to identify the likely non-vegetarians: the ones who eat. The identification isn’t perfect though. Some vegetarians may have started with an empty stomach, and gave in to eating meat, even though in normal situations they would not do so. It could also be the fact that some meat-eaters felt ill that day and didn’t eat anything, when in good health they would eat the meat with alacrity. What you have accomplished is to have taken a sample of both meat-eaters and vegetarians and reduced the sample so that meat-eaters are a larger portion of the sample.
A similar method was employed to reduce the variety of bacteria in the milk samples so that S. aureus would be more prominent. A portion of each milk sample was placed in a petri dish containing a liquid referred to as an agar, which is just a gelatinous substance derived from algae. This particular agar was the Baird-Parker agar, which contained food for certain Staphylococci. However, the agar also contained lithium chloride and tellurite, two substances that inhibit the growth of other microbial types. The petri dish could then be said to have food for some bacteria and poison for others. The colonies able to grow on this agar may contain a number of different bacteria types, and one of these might be S. aureus.
The colonies were then transferred to a different agar referred to as a mannitol salt agar, which performed the same basic function as the Baird-Parker agar. Whatever bacteria thriving after both agars were highly likely to be of the Staphylococci family. But to a scientist, “highly likely” is not good enough. To make absolutely sure the bacteria are S. aureus, their DNA had to be extracted and analyzed to see if it matched the known DNA of the S. aureus strains infecting humans.
Figure 4—S. aureus, growing in a Baird-Parker agar
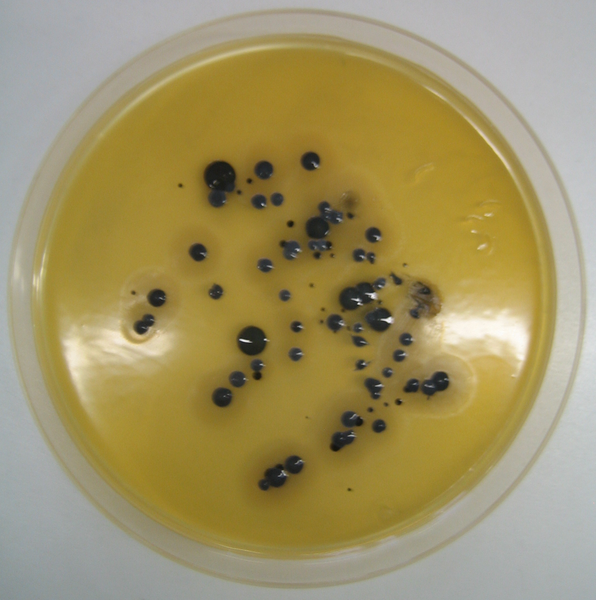
The researchers were not interested in the entire genome of the bacteria though. They knew that if this was S. aureus then there would be one specific segment of the DNA containing the 534 letter sequence below. So they sought to see if its DNA had a segment with this sequence, much like a detective seeks to determine whether fingerprints at a crime scene matched those of a suspect.
Figure 5—Unique 534 letter segment of the S. aureus DNA

Step 3: Copy a portion of the DNA
The DNA of an organism contains the instructions on how it should be built, much like a blueprint of a house. Suppose that we want to take a blueprint for a house and make numerous copies of the master bedroom design. Suppose further we do this by giving a set of tools (e.g., scissors, paper, pencil) to an intern and give her instructions saying she should make a copy of the master bedroom design over and over and over. For the scientist to make many copies of a DNA segment, the scientist must employ nucleotides (the tools), Taq polymerase (the intern), and primers (the instructions).
To understand how the nucleotides, Taq polymerase, and the primers allow one to make multiple copies of the segment of an organism’s DNA, we have to first understand how DNA is built. The twisting ladder of DNA may be among the most recognized shapes today, but most people know little other than its basic shape. The sides of the ladder are made of sugar and phosphate molecules. They contain no information, but instead hold four nucleotides constituting the ladder rungs: Guanine, Cytosine, Adenine, Thymine. These are commonly denoted G, C, A, and T, respectively. Regardless of whether we are talking about bacteria, a tree, or a human, all DNA consists of this same structure: sugars, phosphates, and four nucleotides. The only difference is the length of this twisted ladder and the sequence in which the four nucleotides appear.
Figure 6—The structure of DNA
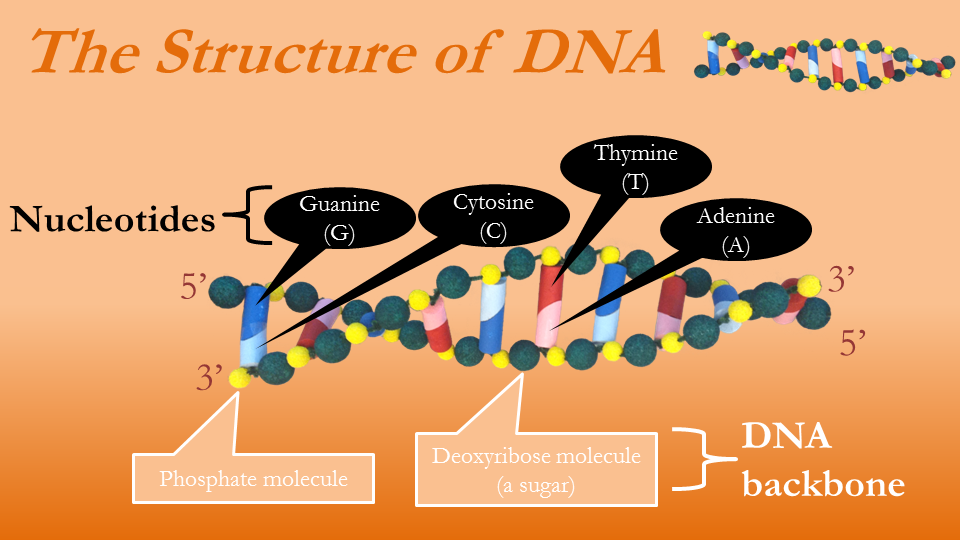
The information within the DNA is coded in the order of the nucleotides. All organisms are built using the sequence of the nucleotides as instructions. The only thing separating humans from earthworms is the size of our DNA and the sequence of the nucleotides. When studying DNA, then, all we need is the sequence of the letters A, C, G, and T. Notice that in Figures 6 and 7 four colors are used to denote four nucleotides, and the rungs of the ladders are really built from two nucleotides. Observe that the nucleotide C is always paired with G, and when A is on one side of the ladder T is on the opposite. This is a universal truth about DNA, true in the genome of the mosquito and the human. This means that once we know one strand of DNA we know the other strand. In Figure 6, the top strand represented in letters would be GACTGACACT, which means the bottom strand must be CTGACTGTGA.
Figure 7—The structure of DNA, a different view
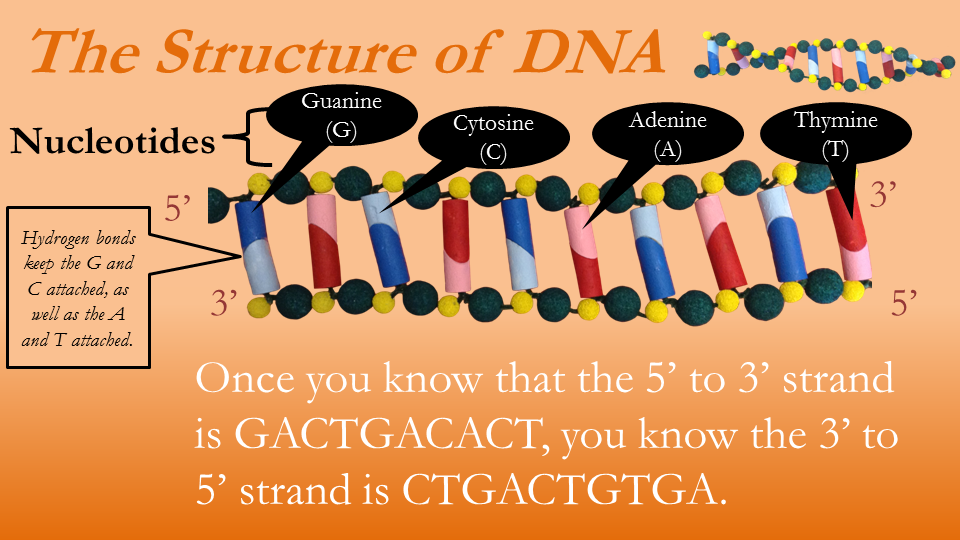
Figure 7 is misleading because there is really no bottom or top strand, as the strands twist about one another, taking turns being on top and bottom. For this reason the sides are instead differentiated by the asymmetry of the carbon atoms comprising the sugar molecules in the DNA spine. One side of sugar molecules will have 5 carbons to the left and 3 to the right, and is thus referred to as the 5' to 3' side.

These simple pictures of DNA do not do justice to the true size of an organism’s genetic complexity. In Figures 6 and 7 each side of the DNA is represented by only 10 letters, whereas the actual genome of S. aureus is over 2.9 million letters, and the genome of a human contains around 3 billion letters! About ten million of the letters in the human genome differ across individuals, giving rise to diverse ethnicities, personalities, and the like. The other 2.99 billion letters in the human genome are virtually identical. You may download a copy of the entire genome of the S. aureus here, but warning, it is over 600 pages long! Rather than downloading it, I suggest you just observe Figure 8, showing a very small portion of the S. aureus DNA.
Figure 8—0.052% of the S. aureus genome

Remember that the DNA is the blueprint of the organism, detailing how it should be built. To see how the sequence of letters in DNA results in actual changes in the organism, consider the simple example of ear wax in humans. There is one specific location in the human genome where either a G or an A will appear. If G appears the person has wet earwax, and if A appears they have dry earwax (if C or T appeared instead it would be a genetic mutation, perhaps resulting in no earwax). That is an overly simplistic example, though. Most genes are not a single letter, but a sequence of letters.
Back to the Matyi study. After using both agars the researcher was ready to inspect the bacteria DNA. To determine whether it was S. aureus the researchers separated the DNA from the rest of the bacteria through the brute force of centrifuges and other machines. A process referred to as polymerase chain reaction (PCR) was then used to make millions of copies from a very specific portion of the DNA. This process is rather complicated, and is best seen in a video tutorial, but a basic description is provided here.
Figure 9—Polymerase Chain Reaction (PCR)

The PCR process basically replicates the DNA in a way such that the vast majority of the replications copy only a very specific portion of the DNA. It would be like a detective collecting a variety of fingerprints but only keeping prints from one particular person. First, the DNA is heated to 203 degrees Fahrenheit, which causes the two strands of a DNA to separate. Then the mixture is cooled to 1400 Fahrenheit, a temperature ideal for DNA replication. We need to bring in our intern, who does the copying. This is the Taq polymerase, which is like a machine that, when primed to land on a piece of DNA, replicates that DNA in one direction from the primer all the way to one end. Some primers copy to the left and some to the right, and when they begin copying, they generally don’t stop until they have reached the DNA’s end. For the new DNA to be built it must have the building blocks, so the researcher adds plenty of A, C, G, and T. Then the primers act as the instructions, telling the polymerase where on the DNA to begin copying.
Figure 10—The steps in PCR

As an example, if one of the separated DNA strands is: TTTAGTCTTCAA, the primer may tell the polymerase to build a new strand starting at the sequence TTT and building to the right (the reader’s right). The polymerase will then place on the primer and every nucleotide to the right its complementary nucleotide. Since the primer is TTT, the polymerase will attach to it AAA. After doing this for the whole strand the polymerase attaches AAATCAGAAGTT to the strand TTTAGTCTTCAA. Notice it doesn’t copy the sequence TTTAGTCTTCAA exactly, but instead builds the nucleotide sequence that would appear on the opposite side of the DNA ladder. Where there is a T, it places an A, where there is a C, it replaces it with a G, and so on.
The primers are simple enzymes chosen such that, after the mixture is heated to 203 and cooled to 140 degrees many times (a process taking only a day or less) what results is a mixture comprised almost exclusively of a very specific part of the DNA. This means that if a strand is randomly sampled from the mixture it will almost certainly be this strand. Then, if this strand is the same size as the strand that would result if the bacteria were indeed S. aureus, the bacteria’s identity is confirmed. The primers used depends on the species and what DNA segment the researcher is targeting. Such primers are easily purchased from companies by telling them the letter sequence of the primers they desire. Dr. Matyi and her colleagues knew what type of primers they would need based on previous studies of S. aureus.
Step 3, again
This is a complex process, and is worth studying in detail, so let’s review it again in a slightly different way, demonstrating what happens after each cycle of heating and cooling.
We start with a strand of DNA, like that below. It is a very long chain, but we are interested in copying only a very small segment of it—the segment enlarged in the figure. Remember that although both the top and bottom sides are different, once you know one you know the other. For these reasons, I refer to both sides as if they are the same identical strand.
The DNA is heated so that the two sides separate, after which the mixture is cooled so that the polymerase can begin buidling. This is the beginning of cycle 1.
Figure 11—Beginning of cycle 1: Separate the DNA using heat

To copy just the targeted strand, the researchers must design two primers. One that will fit on the right side of one strand that will direct the polymerase to copy everything to the left, and another primer that will sit on the left side on the opposite strand and direct the polymerase to copy everything to the right. The primers are designed so that the DNA on and between the two primers is the targeted DNA the researcher wishes to replicate.
All the researchers really need to do is identify a sequence at each end of the targeted DNA and give the information to the company (in a series of letters), which will make a customized primer. For instance, the primers in this example would be CC and TT. Notice that at no other place on the DNA is there a CC on one side or a TT on the other, making the primers fit on one unique place of the DNA.
Figure 12—Parts of the DNA are copied

When the polymerase follows the primers’s instructions it will create two lopsided DNA strands. Both contain the targeted DNA, but one also contains all DNA to the left of the target and another containing all the DNA to the right of the target. There are now four DNA strands: two full strands and two lopsided strands. cycle 1 is completed.
Cycle 2 begins by heating the mixture again so that the DNA separate. Once separated, the temperature is lowered and the Taq polymerase begins copying parts of each DNA strand.
Figure 13—Beginning of Cycle 2

When Cycle 2 is complete it has produced two more lopsided DNA segments and two new strands that precisely equal the targeted DNA.
Figure 14—New DNA strands built in cycle 2

Notice that the number of full DNA strands remain the same, but now there are four lopsided strands and for the first time, two strands which contain only the targeted sequence.
Figure 15—End of cycle 2

As before, cycle 3 begins when the mixture is heated and then cooled for a third time so that the DNA are separated and then copied.
Figure 16—Cycle 3 begins

Again, the number of full strands remain the same, and two more lopsided strands are created. At each cycle of the PCR two more lopsided strands are created but from cycle 2 forward, the number of target strands (containing only the target sequence) more than doubles. When this repeats over and over, the proportion of target strands will approach 100%, making it almost certain that any strand pulled randomly will be that targeted strand.
Figure 17—End of cycle 3

PCR is particularly useful because it doesn’t take many cycles for target strands to dominate the population of DNA strands. To illustrate, suppose that you must reach into a jar of marbles containing equal amounts of white and black balls, and you wish to pull out only a black ball. And it must be done blind-folded. Your chances of success are only 50%. However, if you could put the balls in a much larger container and add thousands of white balls but no additional black balls, after stirring the balls thoroughly it is very unlikely you would randomly retrieve a black ball. That is why PCR is so useful to scientists. Run it for just 25 cycles and you will have 67,108,812 target strands, but only 50 lopsided strands and only 2 full strands of DNA. A DNA randomly chosen from this collection will almost certaintly be the target strand.
Figure 18—At each step of PCR

Step 4: Are the copies the right size?
The specific primers used are such that if the DNA segment copies are indeed from S. aureus, then the segments will be of a very specific size. Of course, scientists cannot measure the segment with a ruler, so they rely on an indirect way of measuring a segment’s length. To understand how it works, imagine yourself pushing a wheelbarrow as fast as you can for ten seconds. The greater the mass of material in the wheelbarrow the shorter distance you can cover. Distance can then measure size. Scientists then “push” this copied DNA segment along a film of gel using electrical currents as force in what is called gel electrophoresis. Like the wheelbarrow metaphor, the further the DNA is pushed, the smaller the DNA segment must be.
Figure 19—A metaphor for gel electrophoresis
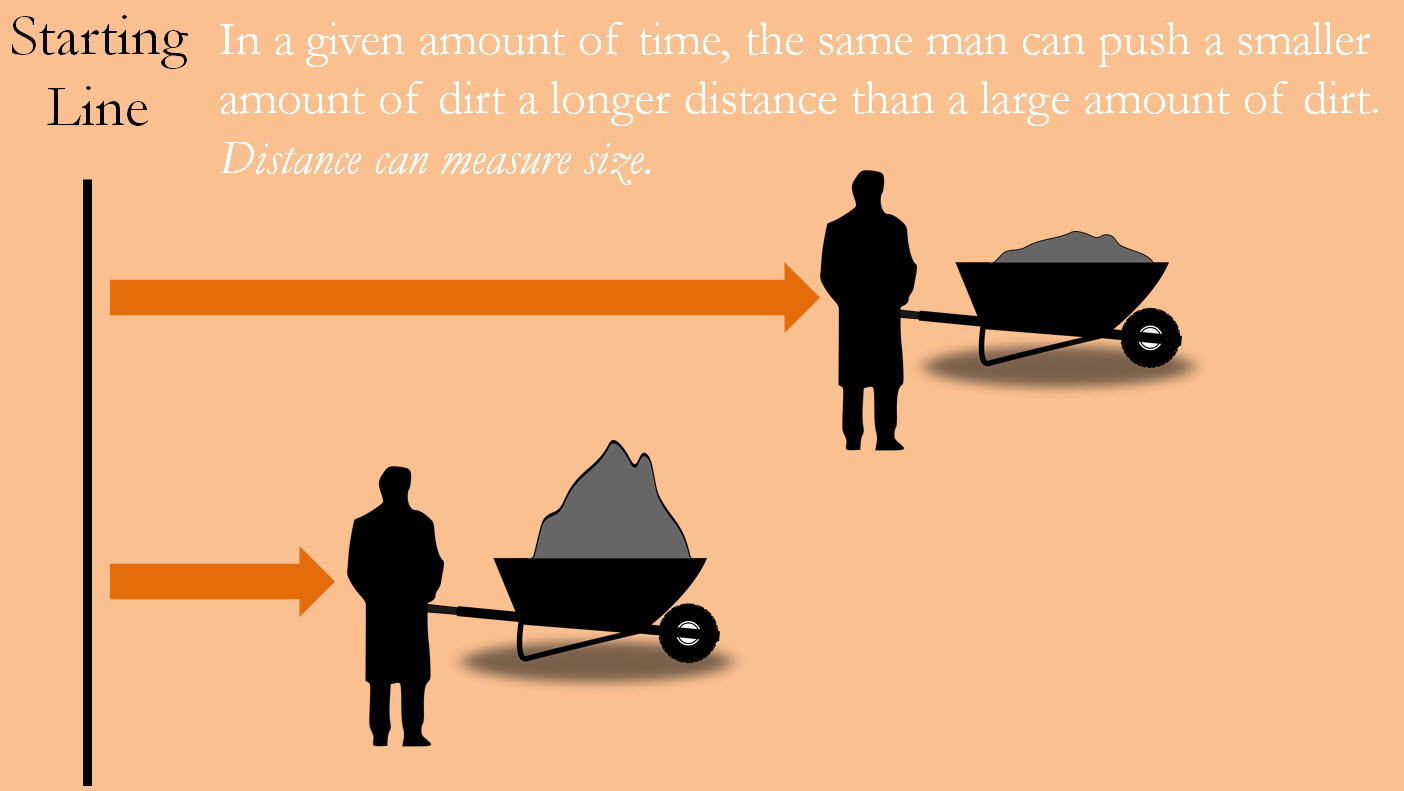
If the targeted DNA segment truly comes from S. aureus it will be a precise length and will cover a precise amount of distance. Since DNA is made up of the acids represented by A, G, C, and T, we can say this segment should have a specific number of letters. If this is the case, then the identity of S. aureus is confirmed.
Figure 20—Overview of DNA identification technique
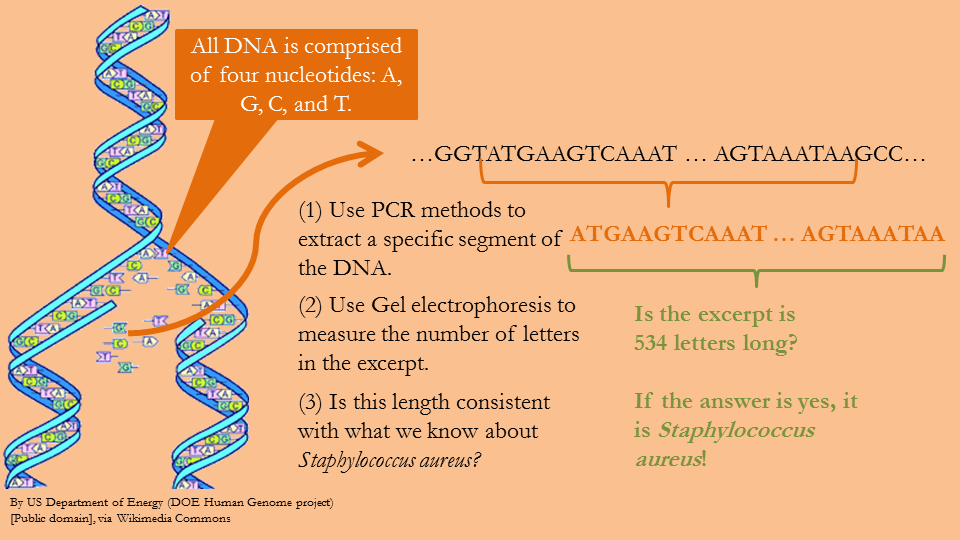
Consider a slightly different overview of the process. The DNA of every organism can be thought of as a string of letters A, G, C, and T. If the bacteria does indeed contain the 534 letter sequence shown in Figure 5, then the PCR process will recognize the segment and will amplify it, which means that in a single day it will make millions of copies of the segment, and each of those segments will be a length of 534 letters. To verify that the DNA segments are indeed 534 letters long, gel electrophoresis is used.
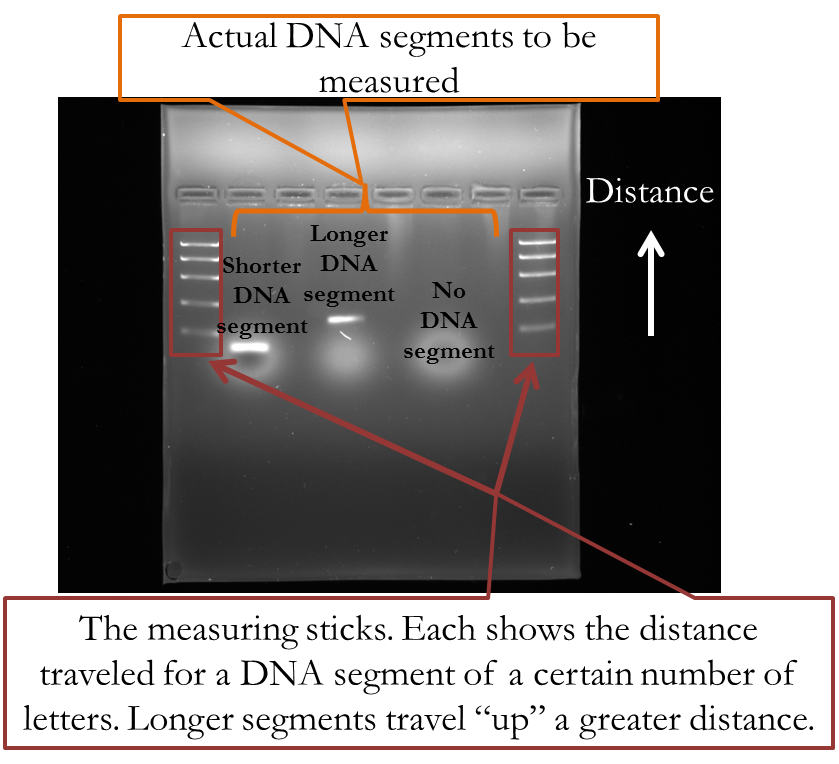
The output of an actual gel electrophoresis is shown below. The series of horizontal bars in the first and last columns are like the measuring sticks. They show the distance segments of a certain number of letters will travel, where the further “up” it travels, the greater the distance. The two horizontal bars in the second and fourth columns are actual DNA segments taken from a PCR process. The segment in the second column contains less letters because it did not travel as great of a distance as the segment in the fourth column. The sixth segment shows what happens if you insert no DNA segment: it doesn’t travel any distance.
When Matyi and her colleagues sought to identify the S. aureus they used gel electrophoresis and determined whether the DNA segment travel the same distance as the measuring sticks said it would if it consisted of 534 letters.
Figure 21—Actual picture of gel electrophoresis results
Results of the Matyi study and antibiotic-resistance of S. aureus.
Results of the Matyi study determined that the same strains of S. aureus that were causing sickness in humans could sicken cattle as well. Both contained that precise DNA segment, like a match of fingerprints. Moreover, it was determined that the S. aureus in the milk samples were resistant to certain bacteria, such that if it infected a person or cow it would be classified as MRSA. Cattle and humans are thus susceptible to the same type of infections with little in the way of medicine to combat it, stressing the interdependence between the health of humans and their livestock.
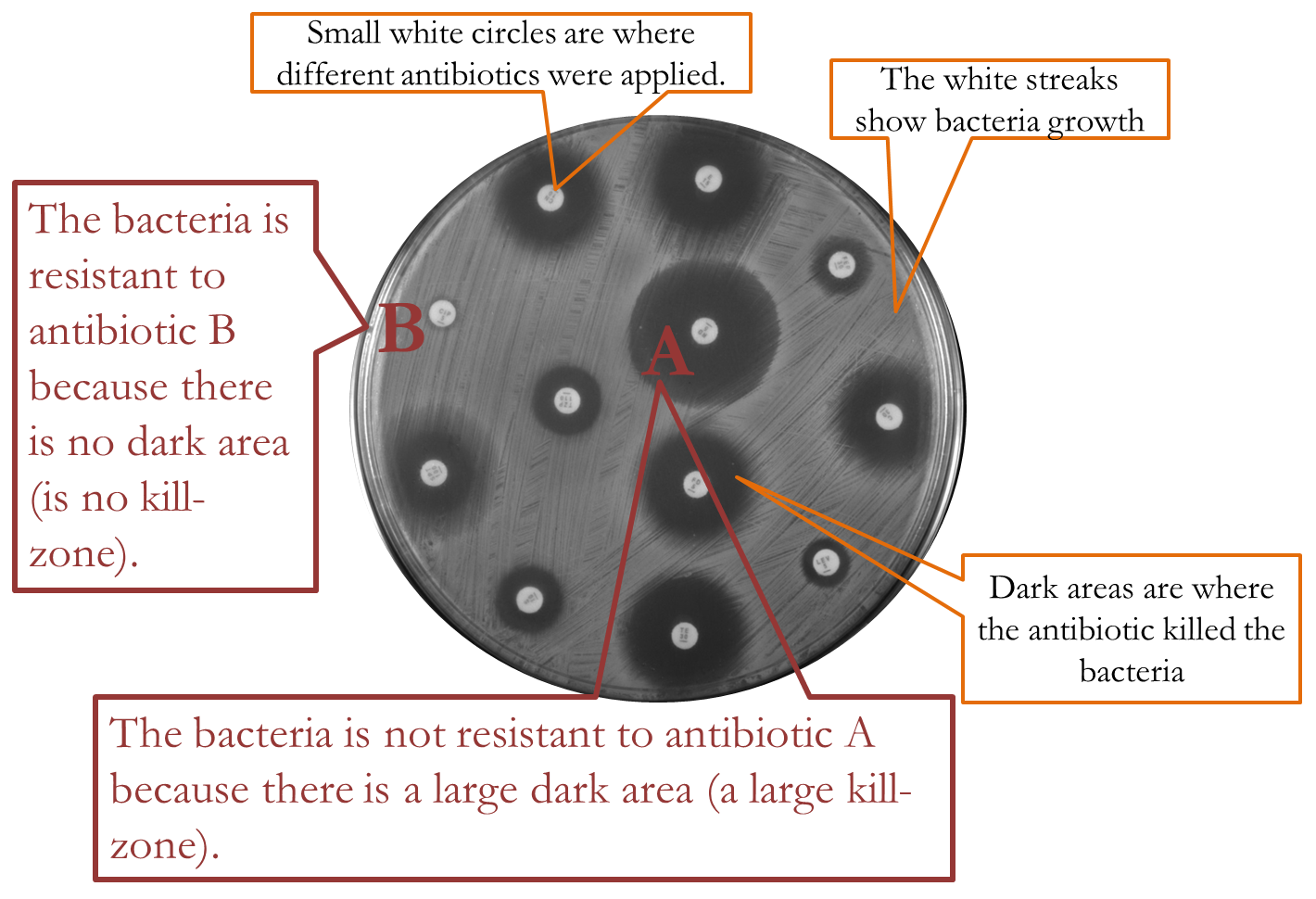
How do researchers determine whether bacteria are resistant to antibiotics? Simple: put some antibiotics on a petri dish of growing bacteria and measure the kill-rate. Below is a picture of an entire petri dish that had been growing bacteria, and the small white circles are where different types of antibiotics were applied. The white streaks are thriving bacteria. The dark circles around the small circles are kill-zones, areas where the antibiotic killed the bacteria. The smaller the dark circle, then, the less effect the antibiotic had on the bacteria, and in the case where there seems no dark circle at all, the bacteria could be said to be resistant to that particular antibiotic. The larger the dark circle the greater the kill-rate, and the greater the antibiotic’s effectiveness in killing the bacteria.
Figure 22—Results of petri dish experiment to determine if a bacterium is resistant to different antibiotics
What this implies is that the way we use antibiotics in livestock can have serious consequences in regards to human health. The opposite is also true. Policies regarding human health cannot ignore the livestock sector, and if we are to reduce our use of antibiotics, both farmers and doctors should cooperate to accomplish a desirable outcome for all.
Towards the future
It will soon be the case that the technology for DNA sequencing, where one gathers information on all the letters constituting the entire DNA of an organism, will become so inexpensive that researchers faced with a similar question as the Matyi study will not use the exact methods described here, but will instead sequence the entire DNA of the bacteria in the milk samples. In the past, sequencing was conducted using the Sanger Method but in the future will probably be done using Ion Torrent technologies.
That doesn’t mean that the information in this article will soon become irrelevant. The Sanger Method employs a number of different tools, one of them being PCR. Although the Ion Torrent technology does not use PCR it does use polymerase to rebuild severed DNA strands.
Understanding PCR is useful for understanding all DNA studies, whatever methods are employed. PCR played a part in beef production when they discovered the gene causing birth defects in one particular Red Angus bull,(A1) and PCR was one of the tools used to identify the gene for tender beef in cattle.(I1) Rice farmers in developing countries now have access to rice seed that will patiently remain dormant during prolonged floods, sprouting when the waters have receded. After identifying the gene sequence that makes some plants possess this trait, they inserted this sequence into more popular rice plants. It took only four years between the time this gene sequence was identified before it was available to farmers, and PCR deserves some of the praise.(E2)
Figures
(1) In public domain. Acquired on May 28, 2014 from Wikimedia Commons at http://en.wikipedia.org/wiki/File:Staphylococcus_aureus_VISA_2.jpg.
(2) By CDC/Bruno Coignard, M.D.; Jeff Hageman, M.H.S. [Public domain], via Wikimedia Commons
(3) By Jóna Þórunn (Own work) [GFDL (http://www.gnu.org/copyleft/fdl.html) or CC-BY-3.0 (http://creativecommons.org/licenses/by/3.0)], via Wikimedia Commons
(4-20) Original figures and/or contain references within the figure.
(21-22) Provided by Stephanie Matyi and the Department of Biochemistry and Molecular Biology.